Zach B. Hancock
@hancockzb.bsky.social
570 followers
260 following
9 posts
Evolutionary biologist, science writer, YouTuber. Assistant Professor at Augusta University. he/him 🏳️🌈
http://www.youtube.com/@talkpopgen
Posts
Media
Videos
Starter Packs
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
Reposted by Zach B. Hancock
C. Brandon Ogbunu
@cbo.bsky.social
· Jul 27
Reposted by Zach B. Hancock
David Gokhman
@david-gokhman.bsky.social
· Jul 26
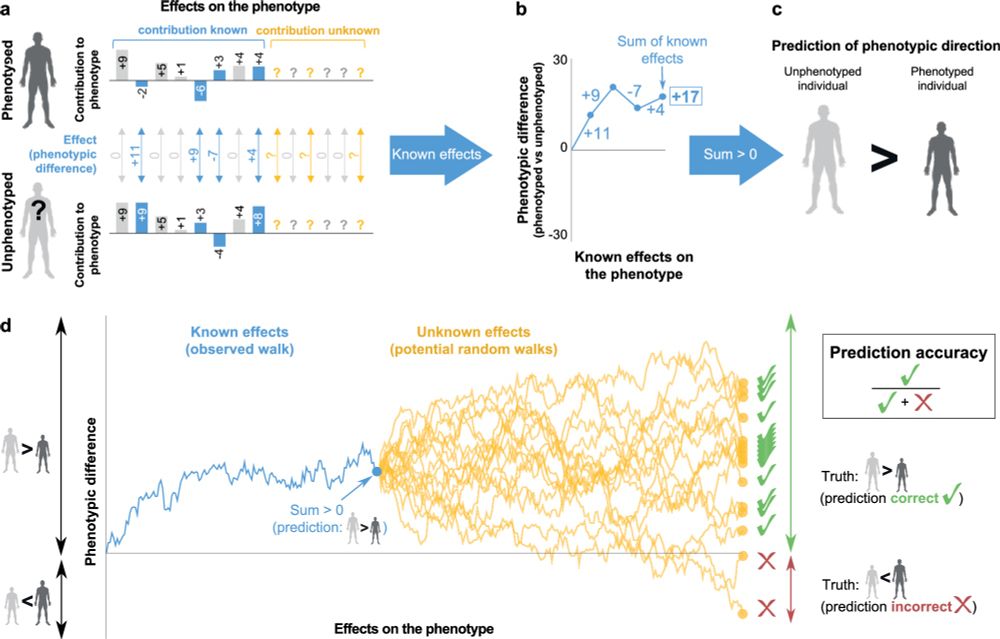
Predicting the direction of phenotypic difference
Nature Communications - Here authors reveal a method to predict key information on phenotypes - their direction. This is achievable even for phenotypes with incomplete genotype-to-phenotype...
rdcu.be
Reposted by Zach B. Hancock
Emily Josephs
@emjo.bsky.social
· Jul 21
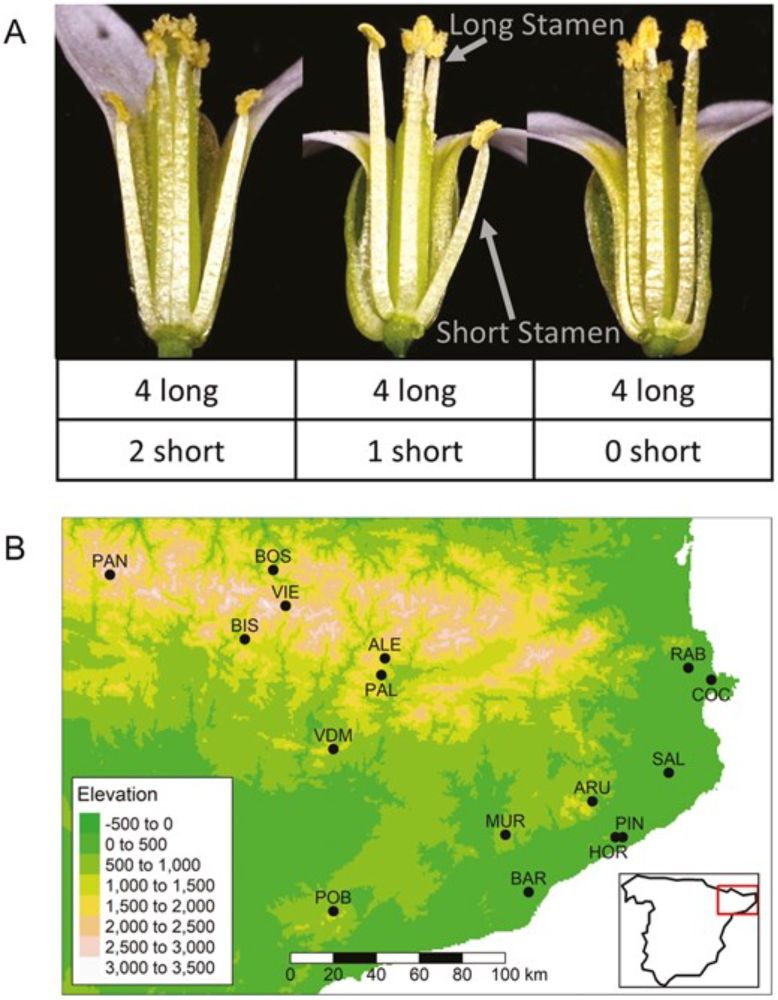
Evaluating the roles of drift and selection in trait loss along an elevational gradient
The evolutionary mechanisms underlying loss or retention of traits that have lost function are poorly understood. Short stamens in Arabidopsis thaliana pro
academic.oup.com