Karim-Jean Armache
@kjarmache.bsky.social
800 followers
130 following
28 posts
GirlDad, Husband, Scientist
Posts
Media
Videos
Starter Packs
Reposted by Karim-Jean Armache
Reposted by Karim-Jean Armache
Reposted by Karim-Jean Armache
Karim-Jean Armache
@kjarmache.bsky.social
· Apr 30
Karim-Jean Armache
@kjarmache.bsky.social
· Mar 11
Karim-Jean Armache
@kjarmache.bsky.social
· Feb 22
Karim-Jean Armache
@kjarmache.bsky.social
· Feb 21
The Groth lab
@grothlab.bsky.social
· Feb 19
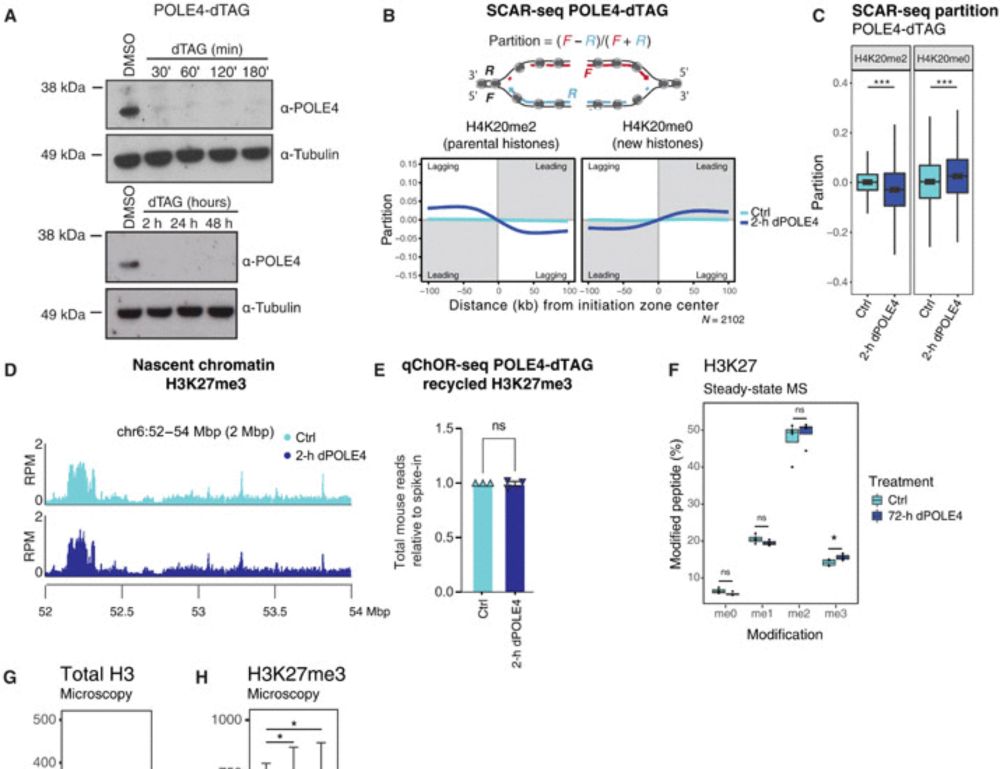
Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability
Losing parental histones during DNA replication fork passage challenges differentiation competence and cell viability.
tinyurl.com
Karim-Jean Armache
@kjarmache.bsky.social
· Feb 15
Karim-Jean Armache
@kjarmache.bsky.social
· Jan 15
Karim-Jean Armache
@kjarmache.bsky.social
· Dec 22
Karim-Jean Armache
@kjarmache.bsky.social
· Dec 13
Reposted by Karim-Jean Armache
Karim-Jean Armache
@kjarmache.bsky.social
· Nov 13
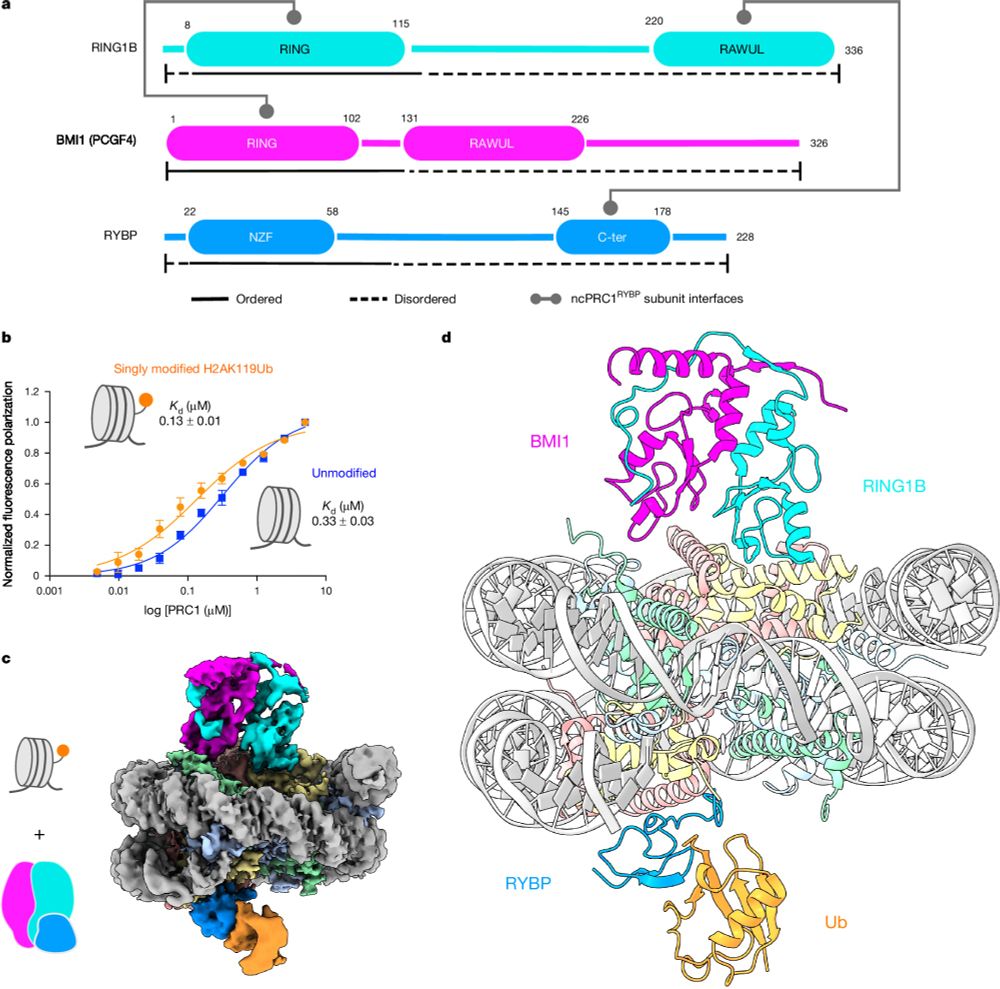
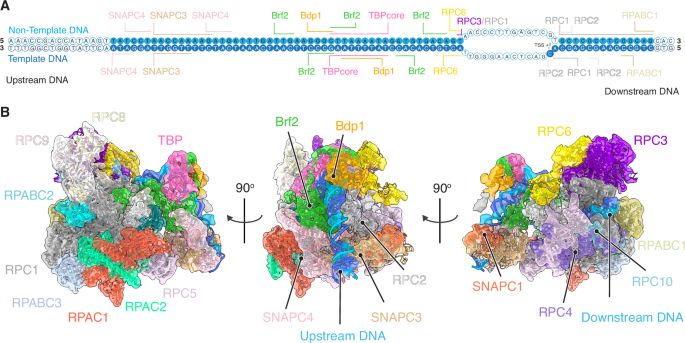
Read–write mechanisms of H2A ubiquitination by Polycomb repressive complex 1
Nature - Cryo-electron microscopy and biochemical studies elucidate the read–write mechanisms of non-canonical PRC1-containing RYBP in histone H2A lysine 119 monoubiquitination and their...
rdcu.be
Karim-Jean Armache
@kjarmache.bsky.social
· Nov 27
Karim-Jean Armache
@kjarmache.bsky.social
· Nov 20
Karim-Jean Armache
@kjarmache.bsky.social
· Nov 18
Karim-Jean Armache
@kjarmache.bsky.social
· Nov 16