Markus Hafner
@markushafnerrna.bsky.social
1.1K followers
920 following
11 posts
Posttranscriptional Gene Regulation @NIH, proud father, 🇩🇪🇺🇸🇭🇷 - any opinions strictly my own.
Posts
Media
Videos
Starter Packs
Reposted by Markus Hafner
Reposted by Markus Hafner
Evelina Tutucci
@anilevetu.bsky.social
· Aug 31

Proximity-specific ribosome profiling reveals the logic of localized mitochondrial translation
Development of LOV-domain-controlled ligase for translation localization (LOCL-TL),
an optogenetic approach for monitoring translation with codon resolution at any defined
subcellular location under p...
www.cell.com
Reposted by Markus Hafner
Reposted by Markus Hafner
Eugene Valkov
@eugenevalkov.bsky.social
· Jul 13
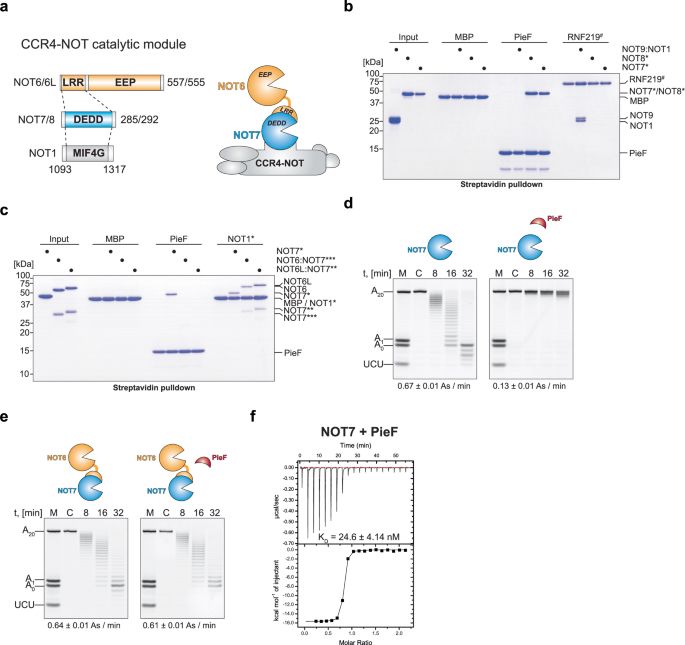
Intracellular pathogen effector reprograms host gene expression by inhibiting mRNA decay - Nature Communications
Intracellular pathogens hijack host gene expression to subvert cellular processes. Here, the authors show that Legionella pneumophila’s PieF effector inhibits the human CCR4-NOT deadenylation machiner...
www.nature.com
Reposted by Markus Hafner
Phil Holliger
@philholliger.bsky.social
· May 29
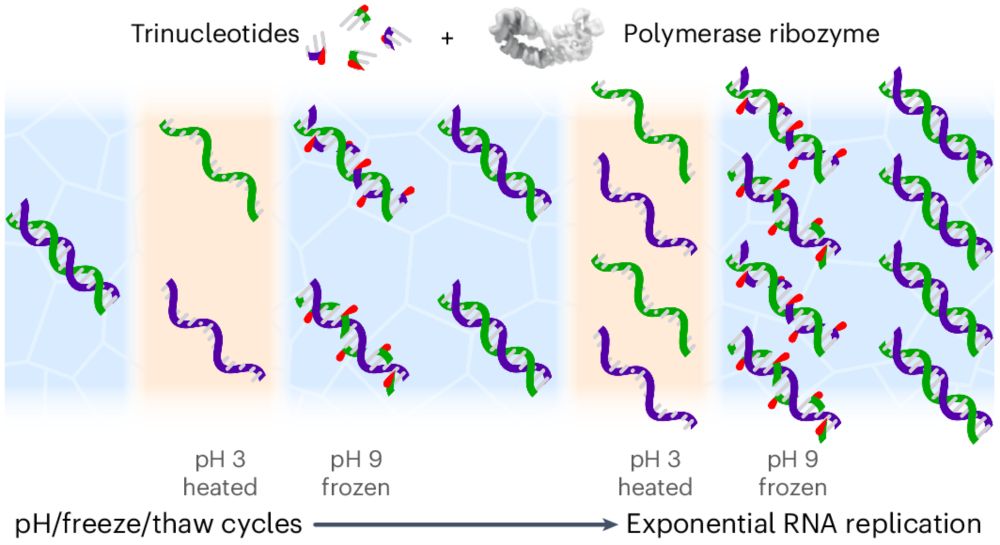
Trinucleotide substrates under pH–freeze–thaw cycles enable open-ended exponential RNA replication by a polymerase ribozyme
Nature Chemistry - Models of abiotic RNA replication suffer from inherent product inhibition arising from the high stability of RNA duplexes. Now, it has been shown that RNA trinucleotide...
rdcu.be
Reposted by Markus Hafner
Anita Corbett
@acorbe2.bsky.social
· May 27
Reposted by Markus Hafner
Liam Faller
@liamfaller.bsky.social
· May 6
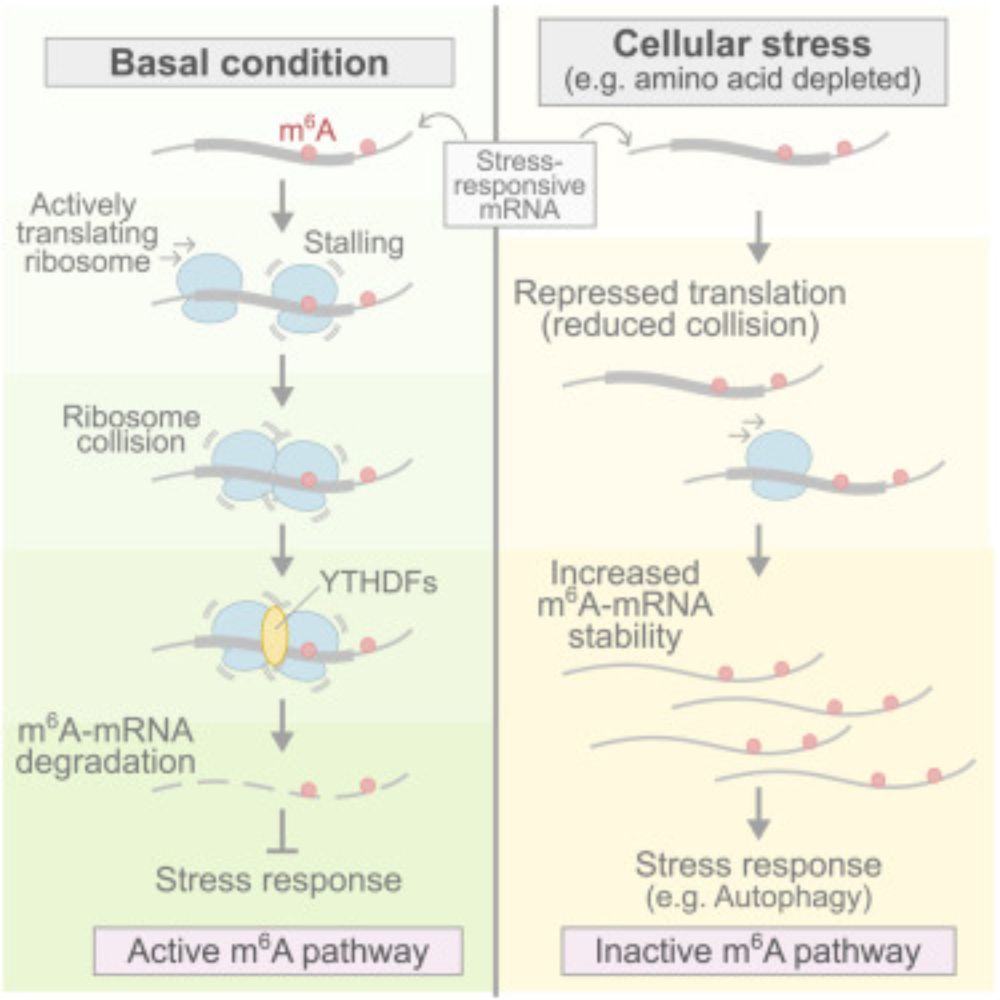
m6A alters ribosome dynamics to initiate mRNA degradation
m6A is a potent inducer of ribosome stalling and collisions that trigger YTHDF-mediated
mRNA degradation, a process that highlights the ribosome as a critical sensor linking
translation dynamics to m6...
www.cell.com
Reposted by Markus Hafner
Reposted by Markus Hafner
Reposted by Markus Hafner
Prajna Hebbar
@prajnahebbar.bsky.social
· Apr 10

Complete sequencing of ape genomes - Nature
Complete sequences of chromosomes telomere-to-telomere from chimpanzee, bonobo, gorilla, Bornean orangutan, Sumatran orangutan and siamang provide a comprehensive and valuable resource for future evol...
www.nature.com
Reposted by Markus Hafner
Reposted by Markus Hafner
Eugene Valkov
@eugenevalkov.bsky.social
· Apr 25
Reposted by Markus Hafner
immunoboys
@immunoboys.bsky.social
· Apr 24

The integrated stress response pathway controls cytokine production in tissue-resident memory CD4+ T cells - Nature Immunology
Panzer and colleagues show that the integrated stress response pathway regulates cytokine translation in CD4+ TRM cells during homeostasis and inflammation.
www.nature.com
Reposted by Markus Hafner
Di Jiang
@dijiang319.bsky.social
· Apr 17
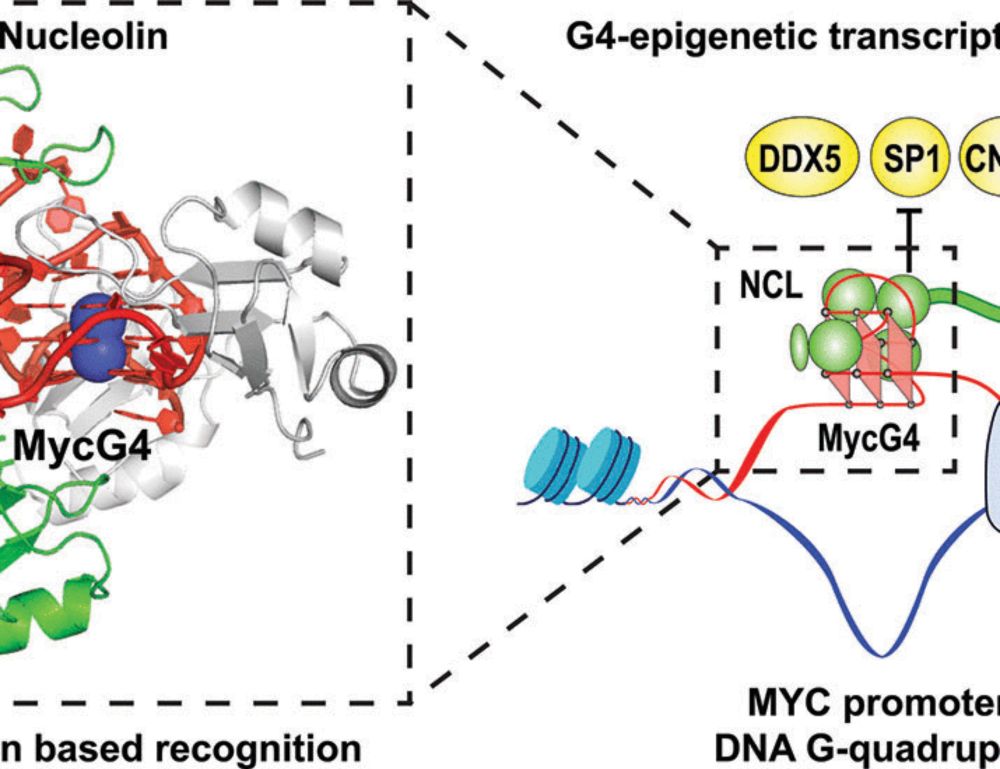
Structural basis for nucleolin recognition of MYC promoter G-quadruplex
The MYC oncogene promoter G-quadruplex (MycG4) regulates transcription and is a prevalent G4 locus in immortal cells. Nucleolin, a major MycG4-binding protein, exhibits greater affinity for MycG4 than...
www.science.org
Reposted by Markus Hafner
Anders H. Lund
@andershlund.bsky.social
· Apr 16
Nature Protocols
@natprot.nature.com
· Apr 16
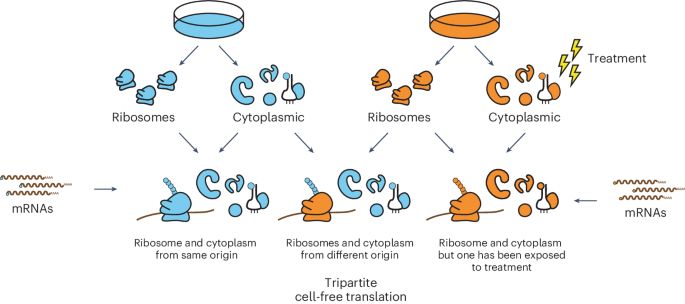
A tripartite cell-free translation system to study mammalian translation
Nature Protocols, Published online: 16 April 2025; doi:10.1038/s41596-025-01155-7This protocol presents a tripartite mammalian cell-free translation system, comprising cytoplasmic and ribosome fractions combined with endogenous or synthetic mRNAs, permitting flexible reconstitution and careful dissection of the mechanisms of translation.
bit.ly
Reposted by Markus Hafner
Nikos Kouvelas
@nickkouvelas.bsky.social
· Apr 17
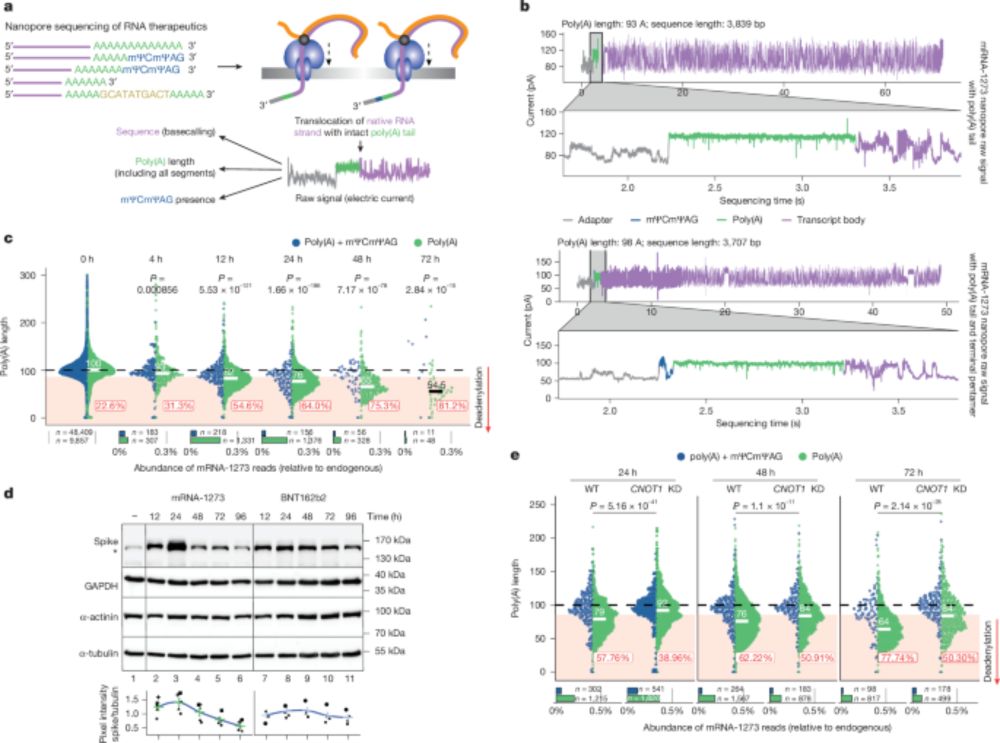
Re-adenylation by TENT5A enhances efficacy of SARS-CoV-2 mRNA vaccines - Nature
Upon intramuscular administration, COVID-19 mRNA vaccines are primarily taken up by macrophages, in which the cellular machinery extends their poly(A) tails, thereby increasing mRNA stability and tran...
www.nature.com
Reposted by Markus Hafner