
Especially interested in the biogenesis of viral proteins.

We show that the transmembrane domain of SARS-CoV-2 Spike protein is not just a passive anchor to the virion surface, but a key player in the viral entry process.
www.nature.com/articles/s41...

www.nature.com/articles/s41...
rdcu.be/eQNsJ

rdcu.be/eQNsJ
We show that the transmembrane domain of SARS-CoV-2 Spike protein is not just a passive anchor to the virion surface, but a key player in the viral entry process.

We show that the transmembrane domain of SARS-CoV-2 Spike protein is not just a passive anchor to the virion surface, but a key player in the viral entry process.

Read more: www2.mrc-lmb.cam.ac.uk/protein-sort...
Read more: www2.mrc-lmb.cam.ac.uk/protein-sort...
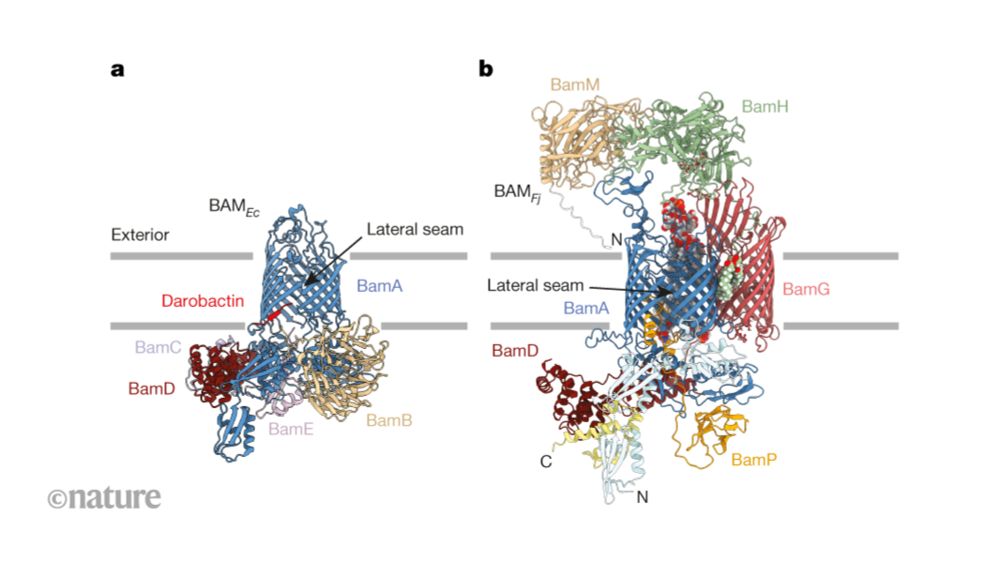
www.nature.com/articles/s41...

www.nature.com/articles/s41...
However, according to reviewers, more evidence is needed on outside-the-groove scramblase activity.
buff.ly/LTxy4yX
However, according to reviewers, more evidence is needed on outside-the-groove scramblase activity.
buff.ly/LTxy4yX
www.nature.com/articles/s41...
www.nature.com/articles/s41...
bit.ly/47b3Mjk

bit.ly/47b3Mjk
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Make my day and check it out!
www.biorxiv.org/content/10.1...
#Cryo-ET #TeamTomo #Cryo-CLEM ❄️🔬❄️🔬❄️🔬❄️🔬
Make my day and check it out!
www.biorxiv.org/content/10.1...
#Cryo-ET #TeamTomo #Cryo-CLEM ❄️🔬❄️🔬❄️🔬❄️🔬
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
Engineering of ATP synthase for enhancement of proton-to-ATP ratio
www.nature.com/articles/s41...
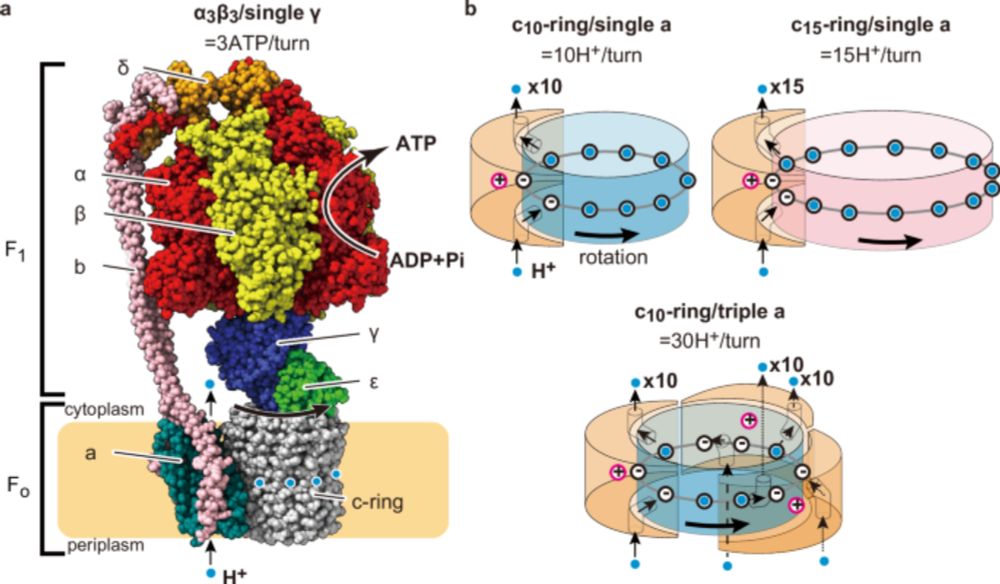
Engineering of ATP synthase for enhancement of proton-to-ATP ratio
www.nature.com/articles/s41...
www.biorxiv.org/node/4580343...
www.biorxiv.org/content/10.1...

www.biorxiv.org/node/4580343...
www.biorxiv.org/content/10.1...
Using U-ExM + in situ cryo-ET, we show how C2CD3 builds an in-to-out radial architecture connecting the distal centriole lumen to its appendages. Great collab with @cellarchlab.com @chgenoud.bsky.social @stearnslab.bsky.social 🙌. #TeamTomo #UExM
www.biorxiv.org/content/10.1...
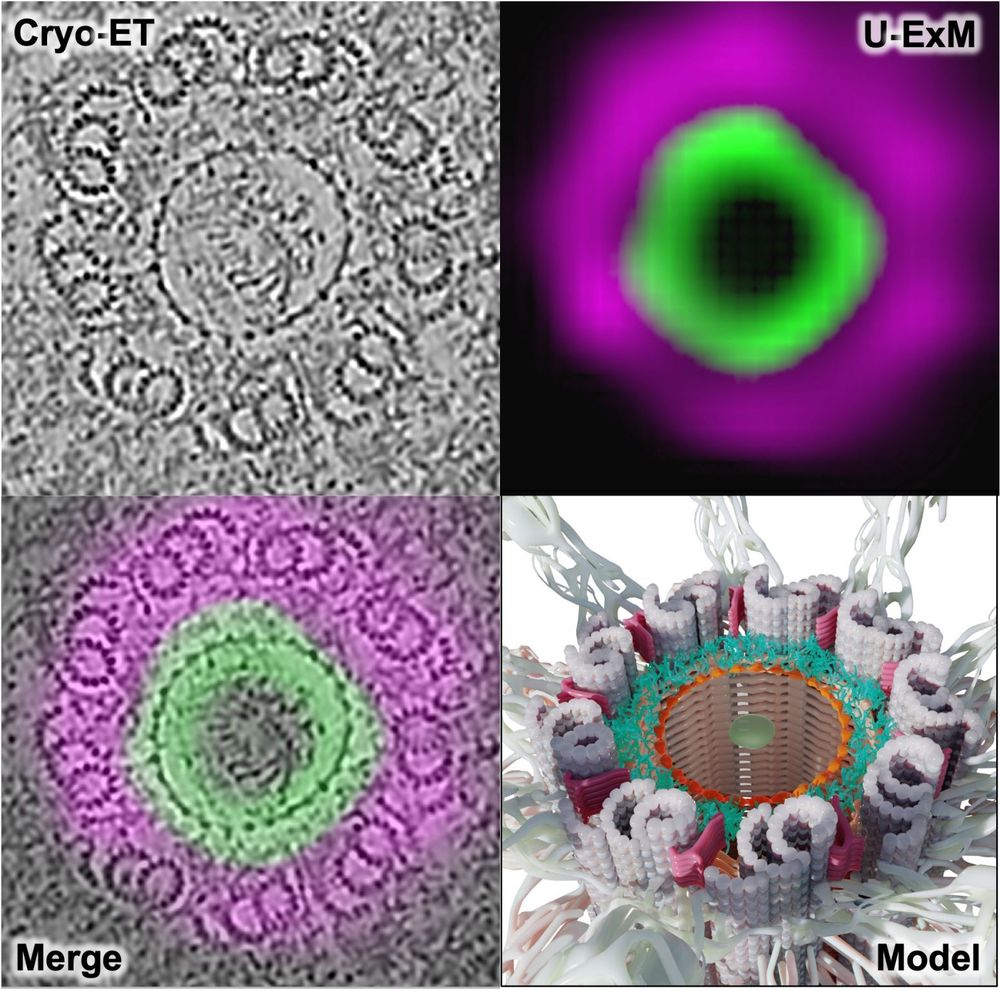
Using U-ExM + in situ cryo-ET, we show how C2CD3 builds an in-to-out radial architecture connecting the distal centriole lumen to its appendages. Great collab with @cellarchlab.com @chgenoud.bsky.social @stearnslab.bsky.social 🙌. #TeamTomo #UExM
www.biorxiv.org/content/10.1...
Our group now call this the “fatty acid hack.”
The #alphafold predictions with and without fatty acids shown below 👇

Our group now call this the “fatty acid hack.”
The #alphafold predictions with and without fatty acids shown below 👇



