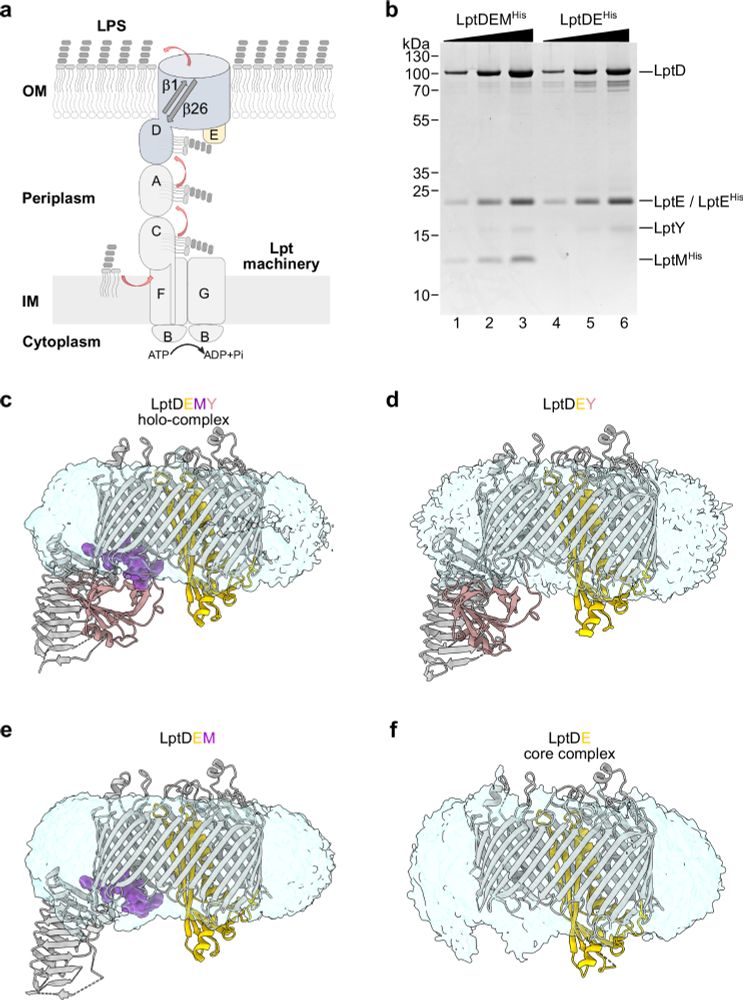
MD simulations of lipids and membrane proteins
Like PIP2, I am negatively charged in the head
www.biorxiv.org/content/10.1101/2025.03.12.642747v1
Shared by @gizmodo.com: buff.ly/yAAHtHq
Shared by @gizmodo.com: buff.ly/yAAHtHq
In recent times these announcements have been around 11am Canberra time. With 2 schemes on the same day, I assume they’ll announce one of them later in the day (probably DECRA first).

In recent times these announcements have been around 11am Canberra time. With 2 schemes on the same day, I assume they’ll announce one of them later in the day (probably DECRA first).


🔗 buff.ly/RFgcZgE
🔗 buff.ly/RFgcZgE
www.nature.com/articles/s41...

#AlphaFold protein and PPI prediction databases
1️⃣ Predictomes (genome maintenance & H2A/H2b) predictomes.org
2️⃣ Flypredictome www.flyrnai.org/tools/fly_pr...
3️⃣ Human interactome prodata.swmed.edu/humanPPI
4️⃣ BFVD (viral proteins) bfvd.steineggerlab.workers.dev
1/2
#AlphaFold protein and PPI prediction databases
1️⃣ Predictomes (genome maintenance & H2A/H2b) predictomes.org
2️⃣ Flypredictome www.flyrnai.org/tools/fly_pr...
3️⃣ Human interactome prodata.swmed.edu/humanPPI
4️⃣ BFVD (viral proteins) bfvd.steineggerlab.workers.dev
1/2

Created for protein–lipid interactions in integral membrane proteins, it also works for peripheral and lipid-anchored proteins too.
pubs.acs.org/doi/full/10....
github.com/keb721/ccd2md

Created for protein–lipid interactions in integral membrane proteins, it also works for peripheral and lipid-anchored proteins too.
pubs.acs.org/doi/full/10....
github.com/keb721/ccd2md


It's *scientific publishing*.
We call this the Drain of Scientific Publishing.
Paper: arxiv.org/abs/2511.04820
Background: doi.org/10.1162/qss_...
Thread @markhanson.fediscience.org.ap.brid.gy 👇

It's *scientific publishing*.
We call this the Drain of Scientific Publishing.
Paper: arxiv.org/abs/2511.04820
Background: doi.org/10.1162/qss_...
Thread @markhanson.fediscience.org.ap.brid.gy 👇

Picked 🔵 because it reminds me of our #ChR2 optotagging experiments 🤓🧪

Picked 🔵 because it reminds me of our #ChR2 optotagging experiments 🤓🧪
We found that BAM in Bacteroides and Porphyromonas gingivalis has a distinct architecture from BAM in Proteobacteria.
doi.org/10.1038/s415...

We found that BAM in Bacteroides and Porphyromonas gingivalis has a distinct architecture from BAM in Proteobacteria.
doi.org/10.1038/s415...
www.canberratimes.com.au/story/907948...

www.canberratimes.com.au/story/907948...



Register here:
us02web.zoom.us/webinar/regi...

Register here:
us02web.zoom.us/webinar/regi...

doi.org/10.1101/2025...
This is work from my PhD in the lab of @rhyswg.bsky.social and many collaborators who contributed amazing data!

doi.org/10.1101/2025...
This is work from my PhD in the lab of @rhyswg.bsky.social and many collaborators who contributed amazing data!

#Biochemistry #CellMetabolism #LipidDroplets

#Biochemistry #CellMetabolism #LipidDroplets

