Conor McClune
@cmcclune.bsky.social
510 followers
930 following
42 posts
Plants, specialized metabolism, synthetic biology, protein evolution, and everything in between.
Postdoc at Sattley & Fordyce Labs, Stanford
Previously SynBio @ Voigt & Laub Labs, MIT
Posts
Media
Videos
Starter Packs
Pinned
Conor McClune
@cmcclune.bsky.social
· Jun 11
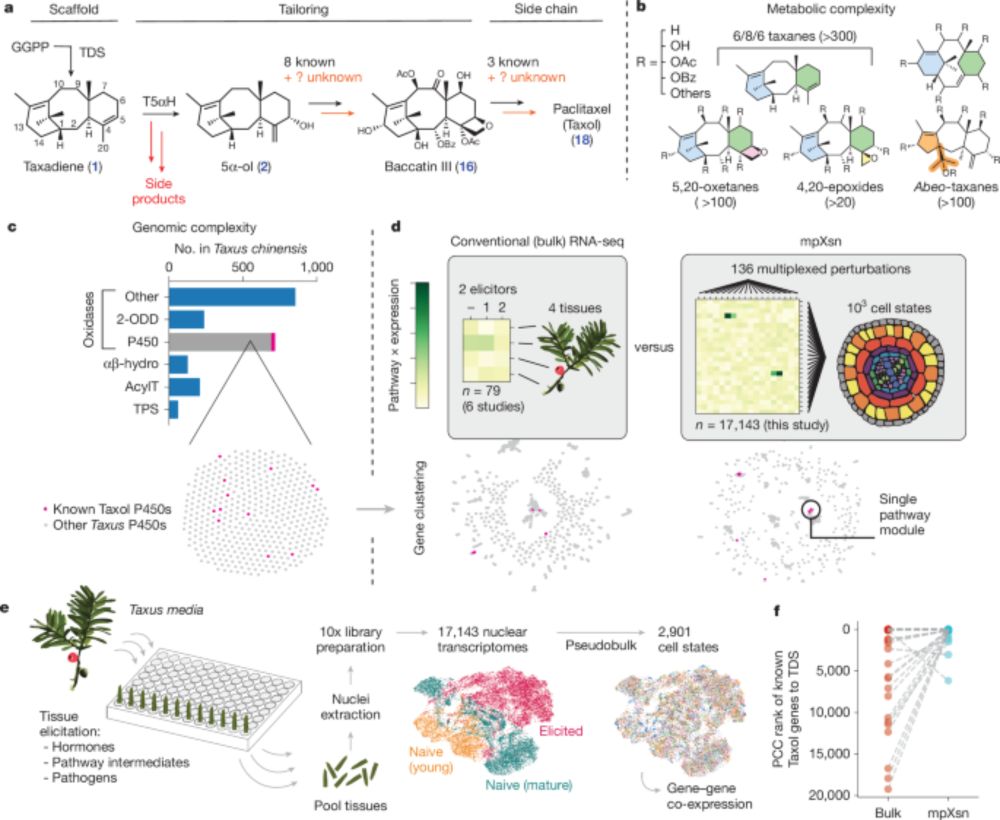
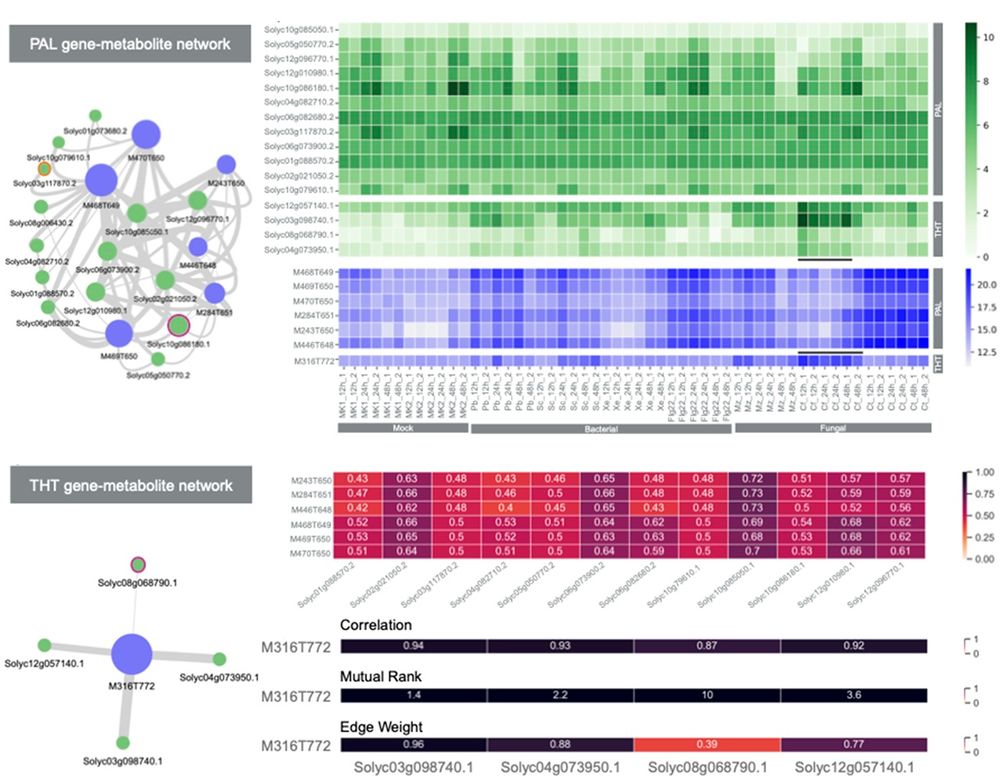
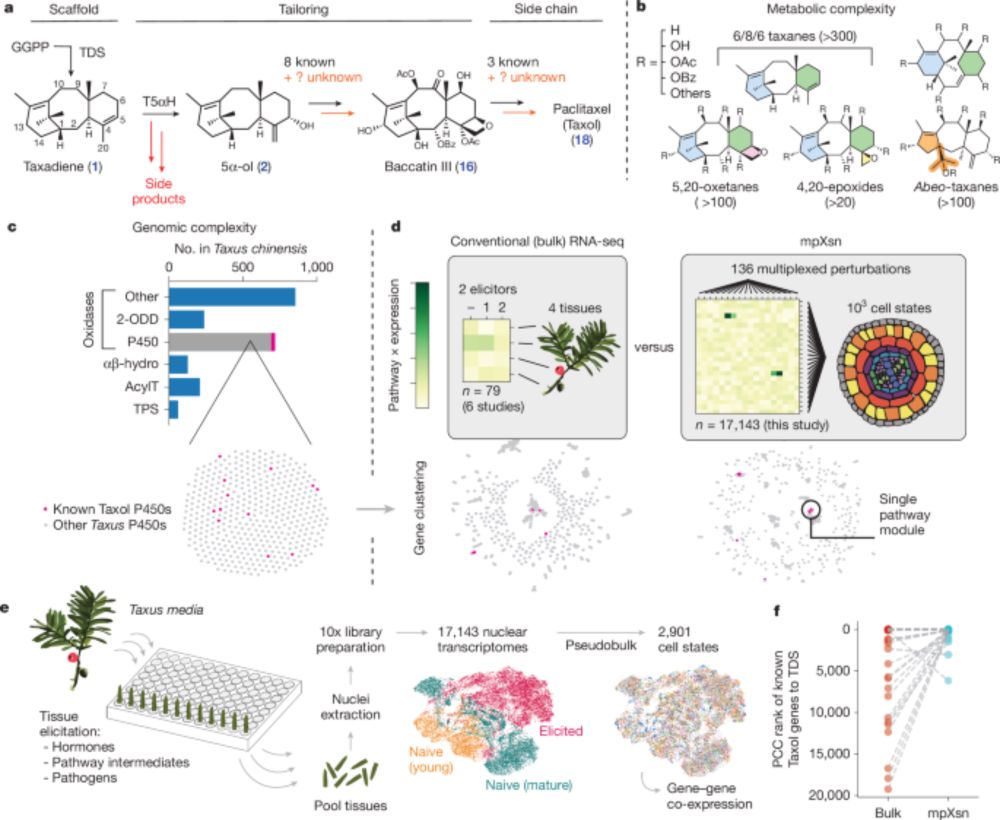
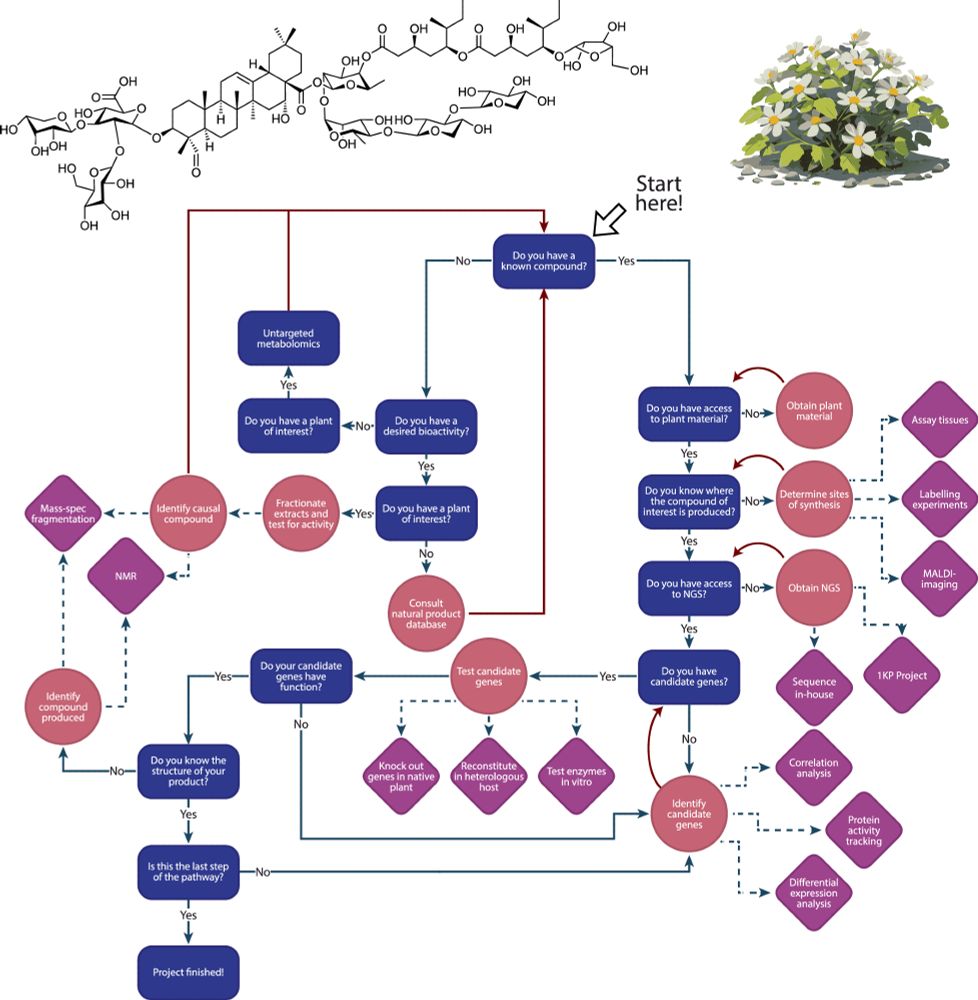
Discovery of FoTO1 and Taxol genes enables biosynthesis of baccatin III - Nature
An approach that combines single-nucleus RNA sequencing and multiplexed perturbation identifies genes that enable the biosynthesis of direct precursors of the anti-cancer drug Taxol, whose curren...
www.nature.com
Reposted by Conor McClune
Reposted by Conor McClune
Maddy Seale
@maddyseale.bsky.social
· Aug 15

Linalool-triggered plant-soil feedback drives defense adaptation in dense maize plantings
High planting density boosts crop yields but also heightens pest and pathogen risks. How plants adapt their defenses under these conditions remains unclear. In this study, we reveal that maize enhance...
www.science.org
Conor McClune
@cmcclune.bsky.social
· Aug 8
Conor McClune
@cmcclune.bsky.social
· Aug 8
Conor McClune
@cmcclune.bsky.social
· Aug 8
Reposted by Conor McClune
SattelyLab
@sattelylab.bsky.social
· Aug 7
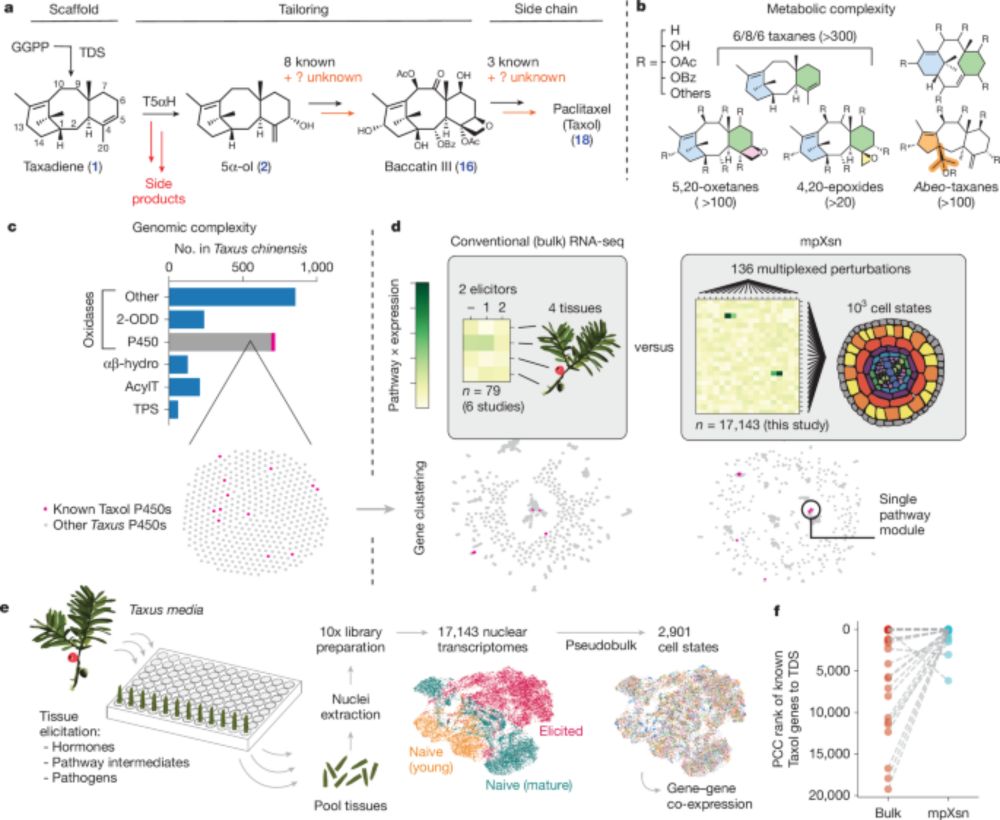
Discovery of FoTO1 and Taxol genes enables biosynthesis of baccatin III - Nature
An approach that combines single-nucleus RNA sequencing and multiplexed perturbation identifies genes that enable the biosynthesis of direct precursors of the anti-cancer drug Taxol, whose curren...
www.nature.com
Reposted by Conor McClune
Reposted by Conor McClune
Reposted by Conor McClune
Conor McClune
@cmcclune.bsky.social
· Jul 11
Reposted by Conor McClune
Conor McClune
@cmcclune.bsky.social
· Jul 11
Reposted by Conor McClune
Quinn Sievers
@quinnsievers.bsky.social
· Jul 11

Discovery of FoTO1 and Taxol genes enables biosynthesis of baccatin III - Nature
An approach that combines single-nucleus RNA sequencing and multiplexed perturbation identifies genes that enable the biosynthesis of direct precursors of the anti-cancer drug Taxol, whose curren...
www.nature.com
Conor McClune
@cmcclune.bsky.social
· Jul 11
Conor McClune
@cmcclune.bsky.social
· Jul 11
Reposted by Conor McClune
Don Moynihan
@donmoyn.bsky.social
· Mar 20
Reposted by Conor McClune
Reposted by Conor McClune
Steven Burgess
@sjb287.bsky.social
· Jul 5
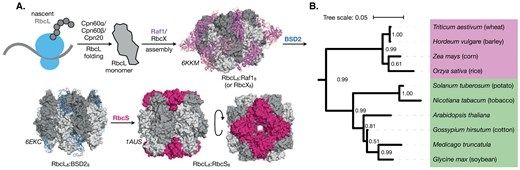
Effects of chaperone selectivity on the assembly of plant Rubisco orthologs in E. coli
We used an E. coli system to examine recognition of plant Rubisco chaperones for different Rubiscos. Identified factors influencing chaperone selectivity e
academic.oup.com
Conor McClune
@cmcclune.bsky.social
· Jul 5
Conor McClune
@cmcclune.bsky.social
· Jul 2