Matthew Clark
@matthew-batisio.bsky.social
1.2K followers
750 following
15 posts
Cellular Structural Biology Wellcome DPhil Student at The University of Oxford.
Currently working in Peijun Zhang’s lab at The Oxford Division of Structural Biology.
Passionate about biomedical animation and science communication: Batisio.co.uk
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Jordan Raff
@jordanraff.bsky.social
· Jul 31
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Reposted by Matthew Clark
Kelly Nguyen
@kellythd-nguyen.bsky.social
· Jul 10
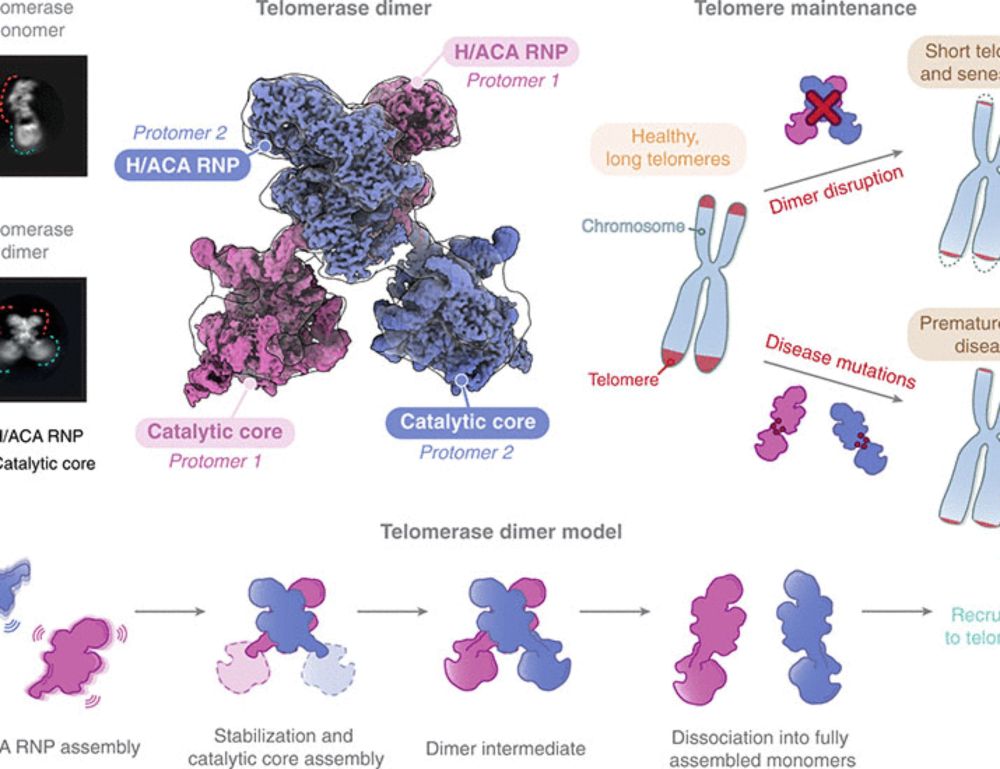
Cryo-EM structure of human telomerase dimer reveals H/ACA RNP-mediated dimerization
Telomerase ribonucleoprotein (RNP) synthesizes telomeric repeats at chromosome ends using a telomerase reverse transcriptase (TERT) and a telomerase RNA (hTR in humans). Previous structural work showe...
www.science.org
Reposted by Matthew Clark
Reposted by Matthew Clark
Susan Schlimpert
@s-lab.bsky.social
· Jul 10

Independent Fellowship in Plant-Associated Microbial Interactions | John Innes Centre
An exciting opportunity for an Independent Fellowship in Plant-Associated Microbial Interactions has arisen at the John Innes Centre. To read the full job description for this role…
www.jic.ac.uk