Kenneth Ehses
@denkenny.bsky.social
410 followers
280 following
6 posts
❄️🔬 cryoEM/ET and subtomogram averaging | Mitochondria are the powerhouse of the cell ⚡️ | PhD Student @uniGoettingen.bsky.social #teamtomo
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Chi-Min Ho
@cmholab.bsky.social
· Aug 18

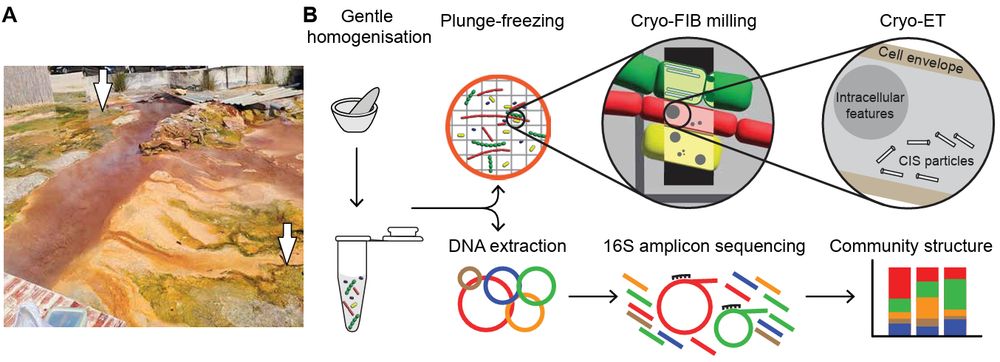
Integrated structural biology of the native malarial translation machinery and its inhibition by an antimalarial drug
Nature Structural & Molecular Biology - Integrated structural biology approach leveraging in situ cryo-electron tomography reveals molecular details of the native malarial translation...
rdcu.be
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
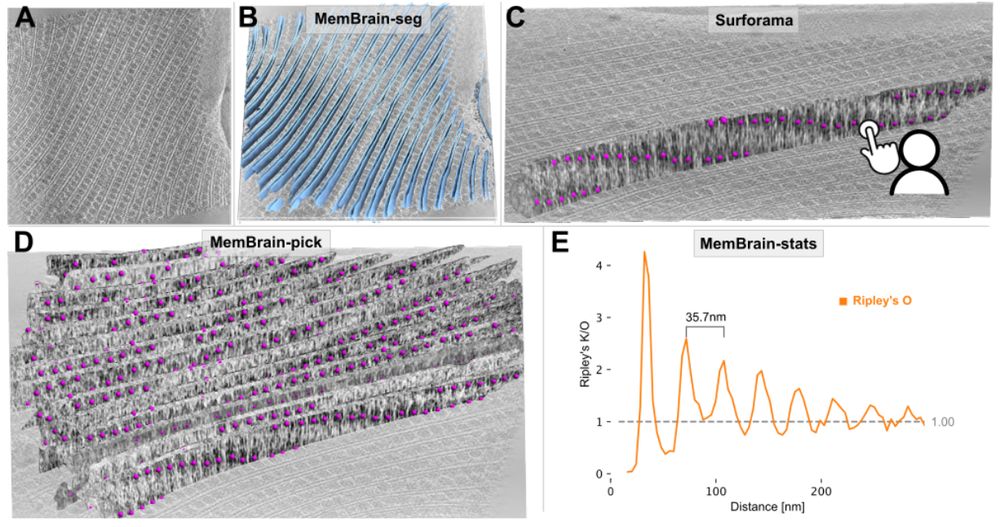
Frank Noe
@franknoe.bsky.social
· Jul 10
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
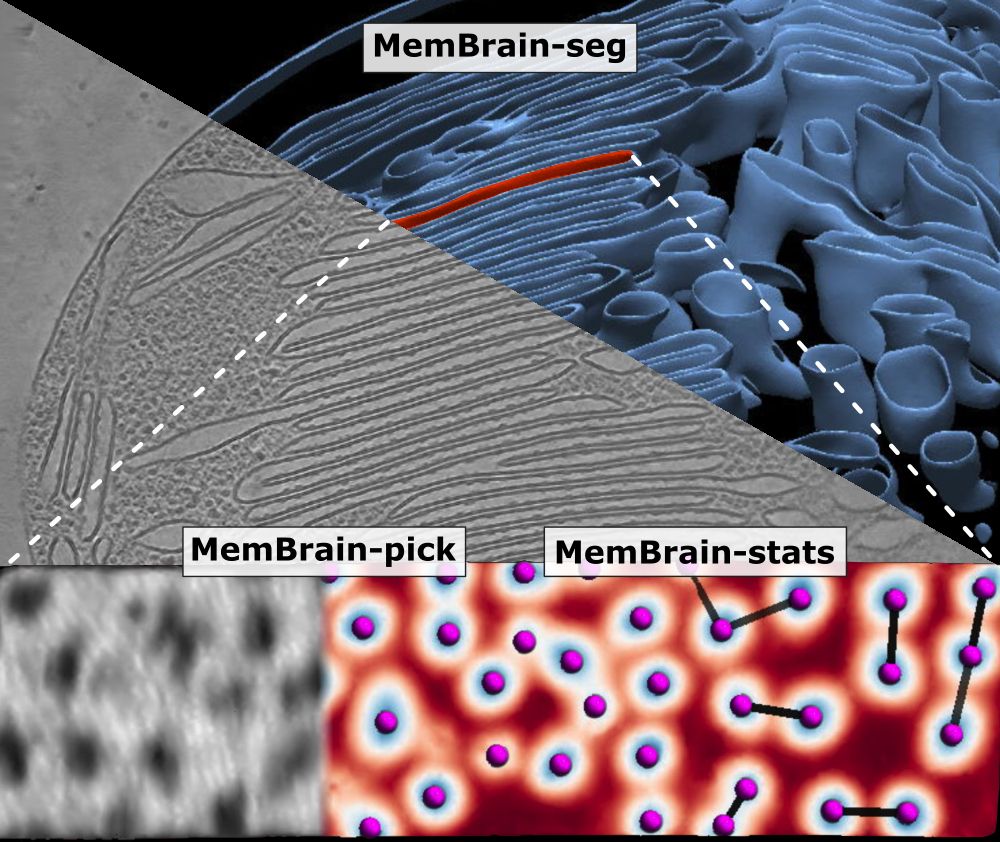
Rasmus Jensen
@rasmusjensen.bsky.social
· May 23
Ewan Birney
@ewanbirney.bsky.social
· May 22

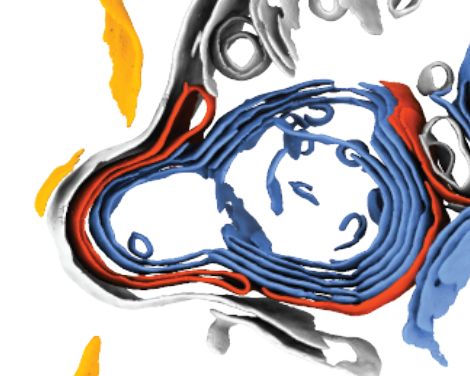
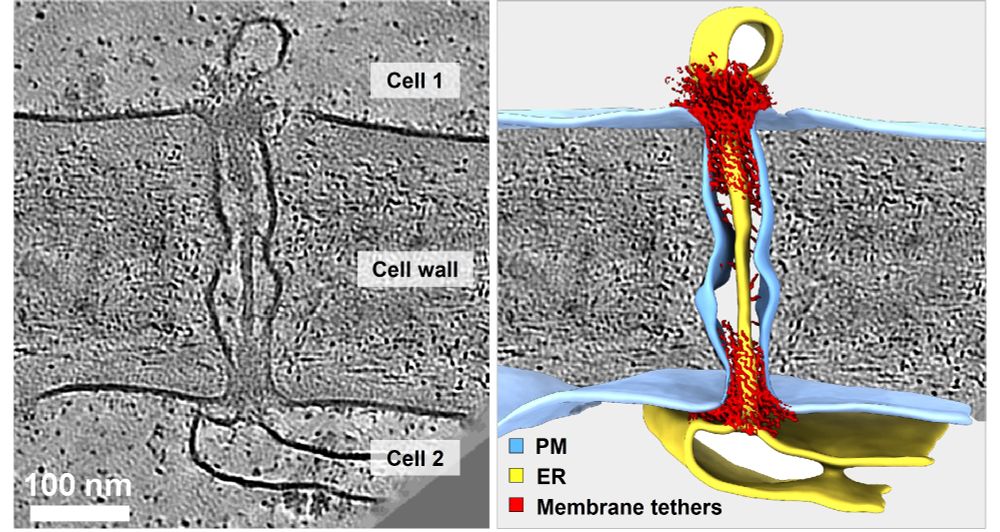
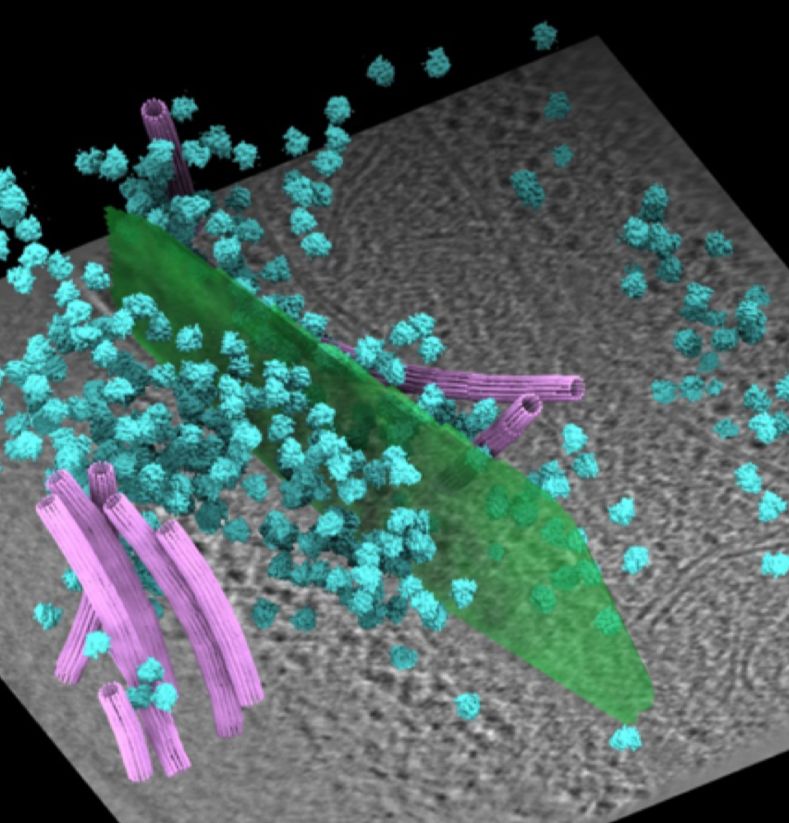
In-cell discovery and characterization of a non-canonical bacterial protein translocation-folding complex
Cryo-electron tomography has emerged as powerful technology for in-cell structural biology, and in combination with breakthroughs in protein structure prediction, offers a unique opportunity for illum...
www.biorxiv.org
Reposted by Kenneth Ehses
Yi-Wei Chang
@yiweichang.bsky.social
· May 15
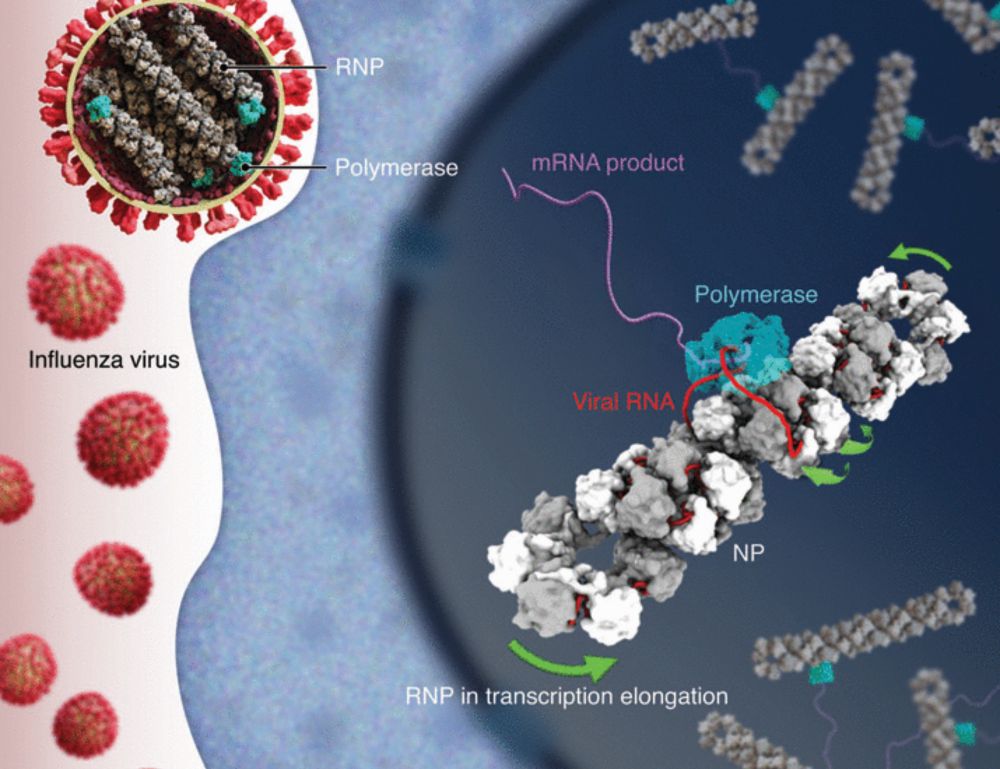
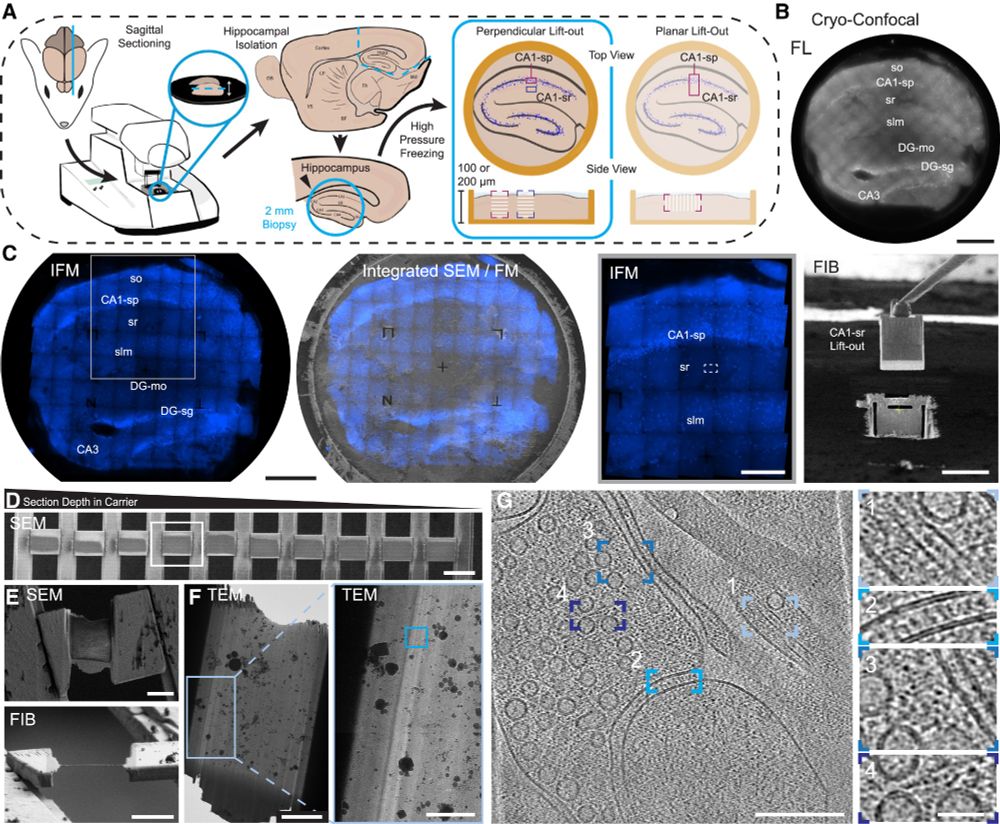
Molecular basis of influenza ribonucleoprotein complex assembly and processive RNA synthesis
Influenza viruses replicate and transcribe their genome in the context of a conserved ribonucleoprotein (RNP) complex. By integrating cryo–electron microscopy single-particle analysis and cryo–electro...
www.science.org
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses
Reposted by Kenneth Ehses